Part:BBa_K3453024
DmMatK Trigger 2.1
This part is a trigger for sequence-based detection of rosewood. It contains a fragment of the MatK gene of Dalbergia maritima var. pubescens (BOLD Acc. n° MADA155-14, MADA063-14, MADA062-14, MADA158-14, MADA065-14, MADA104-14, MADA023-14, MADA071-14, MADA110-14) [1].
Usage and Biology
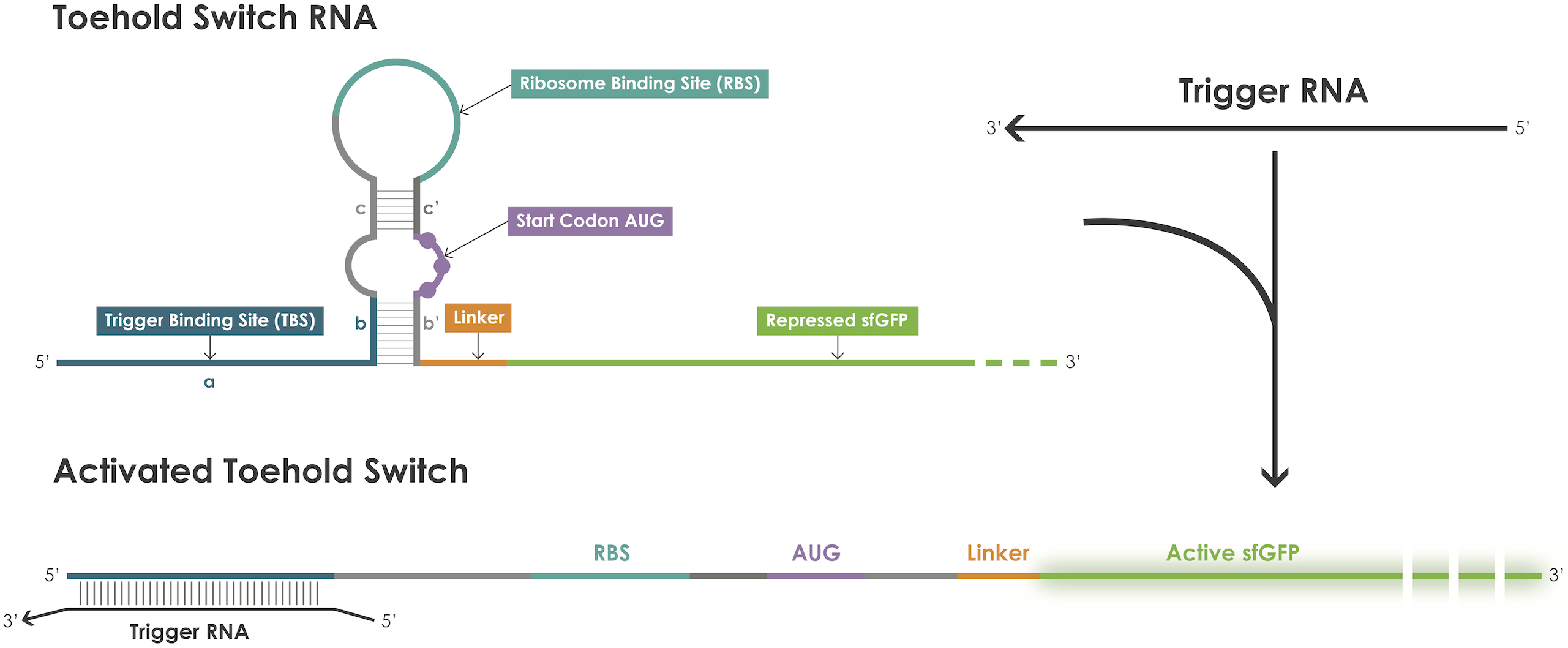
A toehold switch is an RNA–based device containing a ribosome binding site (RBS) and an ATG start codon embedded in a hairpin structure that blocks translation initiation [2]. The hairpin can be unfolded upon binding of a trigger RNA thereby exposing the RBS and the ATG start codon permitting translation of the reporter protein (Figure 1).
Figure 1. Toehold switches principle.
This part is a trigger for the Rosewood DmMatK Toehold Switch 2.1 (BBa_K3453014) and was designed using our internal toehold switch pipeline which calls NUPACK [3] and RBS Calculator for switch prediction and ranking [4].
The functionality of this part was tested under controlled of the T7 promoter (BBa_K2150031) and the strong SBa_000587 synthetic terminator (BBa_K3453000) in the composite part BBa_K3453124.
This part proved to be functional: the transcribed rosewood RNA was able to bind to the corresponding toehold switch and a readable output was generated.
Full results are available on the BBa_K3453114 page in the registry.
References
[1] Hassold S, Lowry PP 2nd, Bauert MR, Razafintsalama A, Ramamonjisoa L, Widmer A. DNA barcoding of Malagasy rosewoods: towards a molecular identification of CITES-listed Dalbergia species. PLoS One (2016) 11, e0157881.
[2] Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell (2014) 159, 925-939.
[3] Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: Analysis and design of nucleic acid systems. Journal of Computational Chemistry (2011) 32, 170–173.
[4] Salis HM. The ribosome binding site calculator. Methods in Enzymology (2011) 498, 19–42.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 26
- 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 26
- 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 26
Illegal BamHI site found at 37 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 26
- 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 26
- 1000COMPATIBLE WITH RFC[1000]
| None |